Technology and Tools
We work on technologies which will allow us to better understand GPCR function. As a lab, we have often found ourselves agreeing with the quotation ascribed to Sydney Brenner: “Progress in science depends on new techniques, new discoveries and new ideas, probably in that order.” (1)
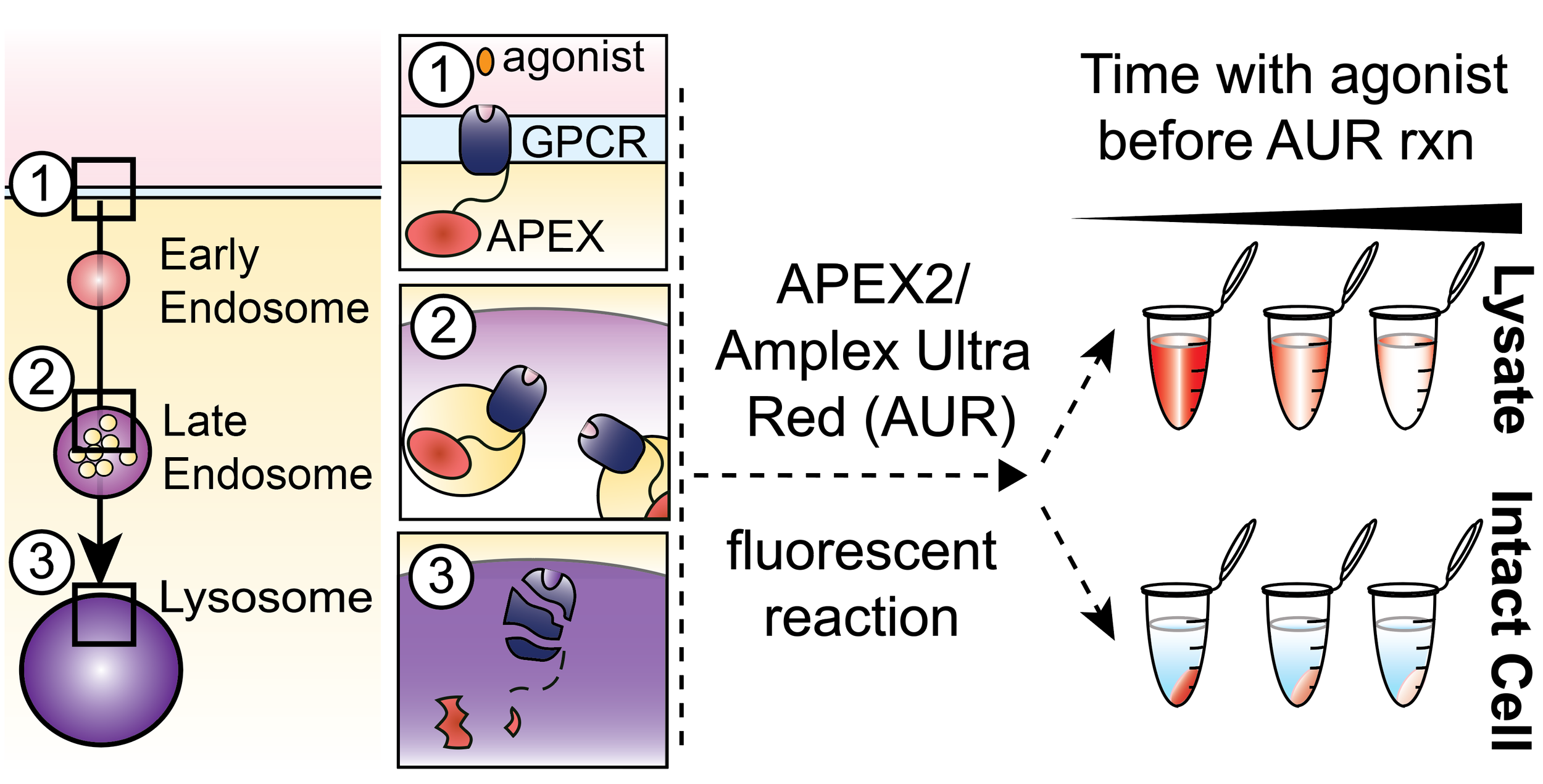
Our current toolkit includes chemical biology methods for GPCR-specific fluorescent cell tagging, proximity labeling, and reporters for GPCR expression and trafficking.